[Spark] 常見錯誤 NoSuchMethodError/ClassNotFound
檸檬爸在開發 Spark, Java, Scala 程式的時候很常遇到 NoSuchMethodError/ClassNotFound 這兩個錯誤,通常出現這兩個錯誤訊息的時候,主要原因是因為 Java Package 的 Dependency Conflict,在開發 Spark 的應用的時候究竟要怎麽去處理會比較好?本篇想要紀錄幾個常用的解法,包含Java 指令, JD-GUI 與 Maven Dependency:Tree 的介紹。
一般來說常見的 NoSuchMethodError 或是 ClassNotFound Error 會如下圖所示,如果是 ClassNotFoundError 可能相對比較好處理,只要加上相對應的 jar 就可以解決,以下我們透過這的範例介紹應該要怎麼 Debug 這兩個常見錯誤。
22/04/14 01:35:36.894 [task-result-getter-1] WARN o.a.spark.scheduler.TaskSetManager - Lost task 0.0 in stage 11.0 (TID 156) (10.0.0.4 executor 0): java.lang.NoSuchMethodError: org.apache.parquet.hadoop.util.HadoopInputFile.getPath()Lorg/apache/hadoop/fs/Path;
at org.apache.spark.sql.execution.datasources.parquet.ParquetFooterReader.readFooter(ParquetFooterReader.java:50)
at org.apache.spark.sql.execution.datasources.parquet.ParquetFooterReader.readFooter(ParquetFooterReader.java:39)
at org.apache.spark.sql.execution.datasources.parquet.ParquetFileFormat.footerFileMetaData$lzycompute$1(ParquetFileFormat.scala:268)
at org.apache.spark.sql.execution.datasources.parquet.ParquetFileFormat.footerFileMetaData$1(ParquetFileFormat.scala:267)
at org.apache.spark.sql.execution.datasources.parquet.ParquetFileFormat.$anonfun$buildReaderWithPartitionValues$2(ParquetFileFormat.scala:270)
at org.apache.spark.sql.execution.datasources.FileScanRDD$anon$1.org$apache$spark$sql$execution$datasources$FileScanRDD$anon$readCurrentFile(FileScanRDD.scala:127)
at org.apache.spark.sql.execution.datasources.FileScanRDD$anon$1.nextIterator(FileScanRDD.scala:187)
at org.apache.spark.sql.execution.datasources.FileScanRDD$anon$1.hasNext(FileScanRDD.scala:104)
at org.apache.spark.sql.execution.FileSourceScanExec$anon$1.hasNext(DataSourceScanExec.scala:522)
at org.apache.spark.sql.catalyst.expressions.GeneratedClass$GeneratedIteratorForCodegenStage1.columnartorow_nextBatch_0$(Unknown Source)
at org.apache.spark.sql.catalyst.expressions.GeneratedClass$GeneratedIteratorForCodegenStage1.processNext(Unknown Source)
at org.apache.spark.sql.execution.BufferedRowIterator.hasNext(BufferedRowIterator.java:43)
at org.apache.spark.sql.execution.WholeStageCodegenExec$anon$1.hasNext(WholeStageCodegenExec.scala:759)
at org.apache.spark.sql.execution.SparkPlan.$anonfun$getByteArrayRdd$1(SparkPlan.scala:349)
at org.apache.spark.rdd.RDD.$anonfun$mapPartitionsInternal$2(RDD.scala:898)
at org.apache.spark.rdd.RDD.$anonfun$mapPartitionsInternal$2$adapted(RDD.scala:898)
at org.apache.spark.rdd.MapPartitionsRDD.compute(MapPartitionsRDD.scala:52)
at org.apache.spark.rdd.RDD.computeOrReadCheckpoint(RDD.scala:373)
at org.apache.spark.rdd.RDD.iterator(RDD.scala:337)
at org.apache.spark.scheduler.ResultTask.runTask(ResultTask.scala:90)
at org.apache.spark.scheduler.Task.run(Task.scala:131)
at org.apache.spark.executor.Executor$TaskRunner.$anonfun$run$3(Executor.scala:506)
at org.apache.spark.util.Utils$.tryWithSafeFinally(Utils.scala:1462)
at org.apache.spark.executor.Executor$TaskRunner.run(Executor.scala:509)
at java.util.concurrent.ThreadPoolExecutor.runWorker(ThreadPoolExecutor.java:1149)
at java.util.concurrent.ThreadPoolExecutor$Worker.run(ThreadPoolExecutor.java:624)
at java.lang.Thread.run(Thread.java:748)首先利用 Java -tvf 指令查出關鍵的 Jar 檔
通常出現 NoSuchMethodError 主要是因為使用到的 jar 有依賴的衝突 (Dependency Conflict) 這時候要除錯的難度就會很高,IBM 的網站有提供一個 shell script 的程式碼幫助我們在很多 jar 裡面找到究竟是哪一個 jar 有用到某個 Class 主要就是利用 java -tvf 指令,以下是 shell script 程式:
if [ $# -ne 1 ]
then
echo " "
echo " "
echo "Usage : FindClass.sh <Class Name/Part of ClassName >"
echo " "
exit -1
fi
for i in `find . -name "*.jar"`
do
jar -tvf $i | grep -i $1 && echo $i
done首先先 cd 到 spark 存放 jars 的位置,通常是 /home/spark-current/jars 接著執行以下指令:
root@5ba55ea3962d46d799fa96666151f347000000:/$ cd /home/spark-current/jars
root@5ba55ea3962d46d799fa96666151f347000000:/home/spark-current/jars$ sh FindClass.sh HadoopInputFile
2437 Thu Sep 30 14:16:04 UTC 2021 org/apache/parquet/hadoop/util/HadoopInputFile.class
./parquet-hadoop-1.12.2.jar
2373 Wed Mar 02 11:36:40 UTC 2022 org/apache/parquet/hadoop/util/HadoopInputFile.class
./adam.jar接著利用 JD-GUI 檢查真正需要的 Jar 檔
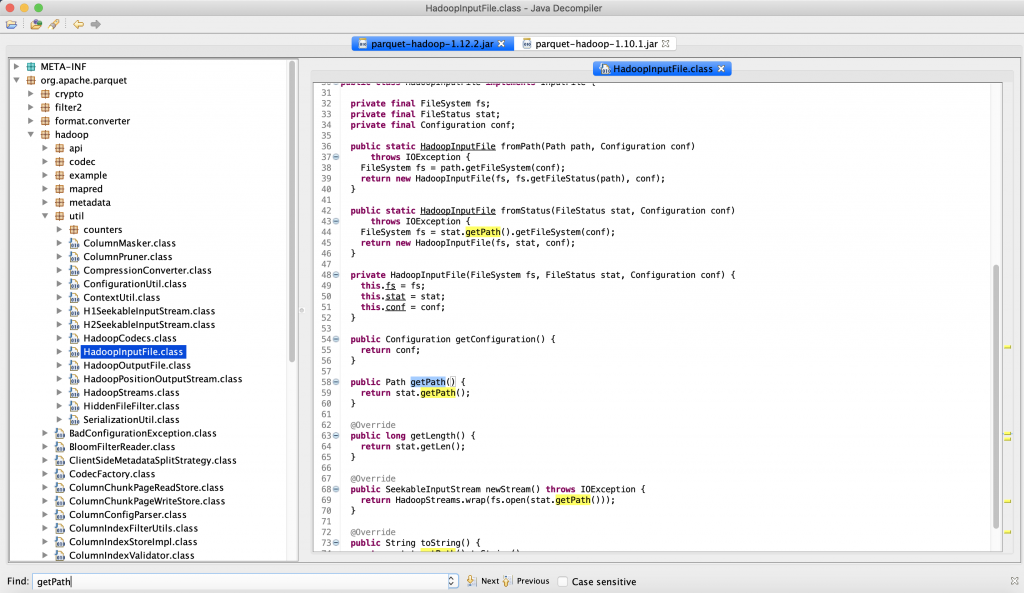
透過第一步的 Java Command 我們知道了 parquet-hadoop-1.12.2.jar 裡面有 HadoopInputFile 這個類別,於是我們利用 JD-GUI 去檢查在 parquet-hadoop-1.12.2.jar 裡面的 HadoopInputFile 是否有 getPath() 這個方法,於下面截圖我們發現 HadoopInputFile 是有 getPath(),因此我們可以推測 pyspark 應該是錯誤拉到了 adam.jar 裡面的 HadoopInputFile 了。
利用 Mvn Dependency:tree 檢查專案中 package 的依賴關係
這邊的 adam.jar 是一個第三方開源的專案,我們利用 Maven Build 之後產生出 jar 檔放在 /home/spark-current/jars 裡面就可以在開發 Spark 程式的時候使用到 Adam 的函式庫。
由於 Adam 是使用 Maven 編譯的,所以我們可以利用 mvn dependency:tree 指令在 pom.xml 得到以下的結果,發現他有使用到 org.apache.parquet:parquet-hadoop:jar:1.10.1:compile,因此研判是 parquet-hadoop.jar 1.10.1 與 1.12.2 發生衝突。
[INFO] org.bdgenomics.adam:adam-assembly-spark3_2.12:jar:LATEST
[INFO] \- org.bdgenomics.adam:adam-cli-spark3_2.12:jar:0.37.0-SNAPSHOT:compile
[INFO] +- org.bdgenomics.utils:utils-misc-spark3_2.12:jar:0.3.0:compile
[INFO] | \- org.scala-lang:scala-library:jar:2.12.10:compile
[INFO] +- org.bdgenomics.utils:utils-io-spark3_2.12:jar:0.3.0:compile
[INFO] | \- org.apache.httpcomponents:httpclient:jar:4.5.7:compile
[INFO] | +- org.apache.httpcomponents:httpcore:jar:4.4.11:compile
[INFO] | +- commons-logging:commons-logging:jar:1.2:compile
[INFO] | \- commons-codec:commons-codec:jar:1.11:compile
[INFO] +- org.bdgenomics.utils:utils-cli-spark3_2.12:jar:0.3.0:compile
[INFO] | +- org.clapper:grizzled-slf4j_2.12:jar:1.3.4:compile
[INFO] | | \- org.slf4j:slf4j-api:jar:1.7.30:compile
[INFO] | \- org.apache.parquet:parquet-avro:jar:1.10.1:compile
[INFO] | +- org.apache.parquet:parquet-column:jar:1.10.1:compile
[INFO] | | +- org.apache.parquet:parquet-common:jar:1.10.1:compile
[INFO] | | \- org.apache.parquet:parquet-encoding:jar:1.10.1:compile
[INFO] | +- org.apache.parquet:parquet-hadoop:jar:1.10.1:compile
[INFO] | | +- org.apache.parquet:parquet-jackson:jar:1.10.1:compile
[INFO] | | \- commons-pool:commons-pool:jar:1.6:compile
[INFO] | \- org.apache.parquet:parquet-format:jar:2.4.0:compile
[INFO] +- org.bdgenomics.bdg-formats:bdg-formats:jar:0.15.0:compile
[INFO] | \- org.apache.avro:avro:jar:1.8.2:compile
[INFO] | +- org.codehaus.jackson:jackson-core-asl:jar:1.9.13:compile
[INFO] | +- org.codehaus.jackson:jackson-mapper-asl:jar:1.9.13:compile
[INFO] | +- com.thoughtworks.paranamer:paranamer:jar:2.8:compile
[INFO] | +- org.xerial.snappy:snappy-java:jar:1.1.1.3:compile
[INFO] | +- org.apache.commons:commons-compress:jar:1.8.1:compile
[INFO] | \- org.tukaani:xz:jar:1.5:compile
[INFO] +- org.bdgenomics.adam:adam-core-spark3_2.12:jar:0.37.0-SNAPSHOT:compile
[INFO] | +- org.bdgenomics.utils:utils-intervalrdd-spark3_2.12:jar:0.3.0:compile
[INFO] | +- com.esotericsoftware.kryo:kryo:jar:2.24.0:compile
[INFO] | | +- com.esotericsoftware.minlog:minlog:jar:1.2:compile
[INFO] | | \- org.objenesis:objenesis:jar:2.1:compile
[INFO] | +- commons-io:commons-io:jar:2.6:compile
[INFO] | +- it.unimi.dsi:fastutil:jar:6.6.5:compile
[INFO] | +- org.seqdoop:hadoop-bam:jar:7.10.0-atgx:compile
[INFO] | | \- com.github.jsr203hadoop:jsr203hadoop:jar:1.0.3:compile
[INFO] | +- com.github.samtools:htsjdk:jar:2.19.0:compile
[INFO] | | +- org.apache.commons:commons-jexl:jar:2.1.1:compile
[INFO] | | \- gov.nih.nlm.ncbi:ngs-java:jar:2.9.0:compile
[INFO] | +- com.google.guava:guava:jar:27.0-jre:compile
[INFO] | | +- com.google.guava:failureaccess:jar:1.0:compile
[INFO] | | +- com.google.guava:listenablefuture:jar:9999.0-empty-to-avoid-conflict-with-guava:compile
[INFO] | | +- org.checkerframework:checker-qual:jar:2.5.2:compile
[INFO] | | +- com.google.errorprone:error_prone_annotations:jar:2.2.0:compile
[INFO] | | +- com.google.j2objc:j2objc-annotations:jar:1.1:compile
[INFO] | | \- org.codehaus.mojo:animal-sniffer-annotations:jar:1.17:compile
[INFO] | \- org.bdgenomics.adam:adam-codegen-spark3_2.12:jar:0.37.0-SNAPSHOT:compile
[INFO] +- org.bdgenomics.adam:adam-apis-spark3_2.12:jar:0.37.0-SNAPSHOT:compile
[INFO] +- args4j:args4j:jar:2.33:compile
[INFO] \- net.codingwell:scala-guice_2.12:jar:4.2.1:compile
[INFO] +- com.google.inject:guice:jar:4.2.0:compile
[INFO] | +- javax.inject:javax.inject:jar:1:compile
[INFO] | \- aopalliance:aopalliance:jar:1.0:compile
[INFO] +- org.scala-lang:scala-reflect:jar:2.12.6:compile
[INFO] \- com.google.code.findbugs:jsr305:jar:1.3.9:compile
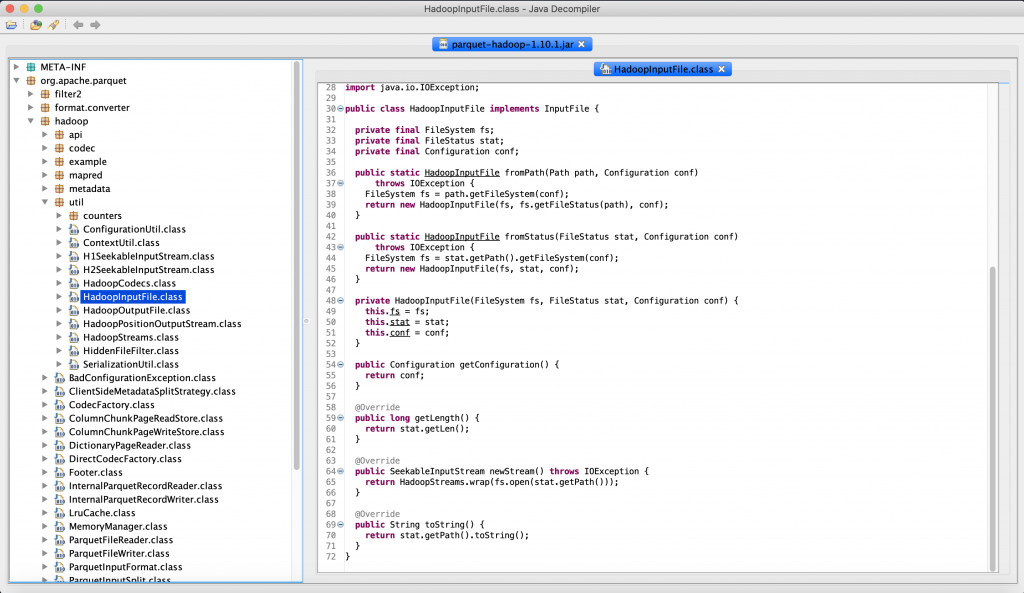
[INFO] ------------------------------------------------------------------------此時進一步用 JD-GUI 確認發現的確是因為 parquet-hadoop-1.10.1.jar 裡面的 HadoopInputFile 沒有 getPath() 的 Method,此時有兩個選擇,升級 adam 支援 1.12.1 或是將 parquet-hadoop-1.12.2 降回到 1.10.1 這兩個方法都可以。
最後希望以上的經驗分享有幫助被這個問題困住的網友。
1. [Spark] 建置自己的 Spark History Server
2. 安裝 Spark 到遠端伺服器
3. [Hadoop] Hdfs Data Integrity with Checksum
